Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling. - Abstract - Europe PMC

A structural model of MurJ. The front view of the model structure of... | Download Scientific Diagram

Overview of the main processes modulated by tagatose in Phytophthora... | Download Scientific Diagram

Loss of specificity variants of WzxC suggest that substrate recognition is coupled with transporter opening in MOP‐family flippases - Sham - 2018 - Molecular Microbiology - Wiley Online Library
Supplemental Figure S3. Enriched transporter families in the six gene expression clusters. (A) Enriched transporter families in

Loss of specificity variants of WzxC suggest that substrate recognition is coupled with transporter opening in MOP‐family flippases - Sham - 2018 - Molecular Microbiology - Wiley Online Library

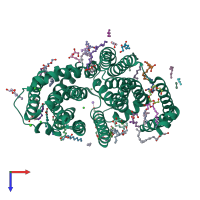
Crystal structure of the lipid flippase MurJ in a “squeezed” form distinct from its inward- and outward-facing forms - ScienceDirect

Crystal structure of the MOP flippase MurJ in an inward-facing conformation | Nature Structural & Molecular Biology

Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling | Journal of the American Chemical Society

Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling | Journal of the American Chemical Society

Crystal structure of the MOP flippase MurJ in an inward-facing conformation | Nature Structural & Molecular Biology
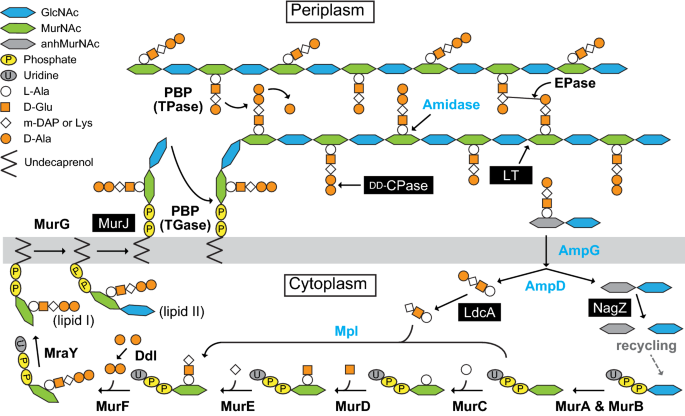
![PDF] Lipid Flippases for Bacterial Peptidoglycan Biosynthesis | Semantic Scholar PDF] Lipid Flippases for Bacterial Peptidoglycan Biosynthesis | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/8da4c1fa261b3e83fa8dc15087f578bf8983ecef/4-Figure3-1.png)
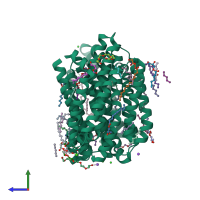


![PDF] Lipid Flippases for Bacterial Peptidoglycan Biosynthesis | Semantic Scholar PDF] Lipid Flippases for Bacterial Peptidoglycan Biosynthesis | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/8da4c1fa261b3e83fa8dc15087f578bf8983ecef/2-Figure1-1.png)




![PDF] Lipid Flippases for Bacterial Peptidoglycan Biosynthesis | Semantic Scholar PDF] Lipid Flippases for Bacterial Peptidoglycan Biosynthesis | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/8da4c1fa261b3e83fa8dc15087f578bf8983ecef/6-Figure4-1.png)