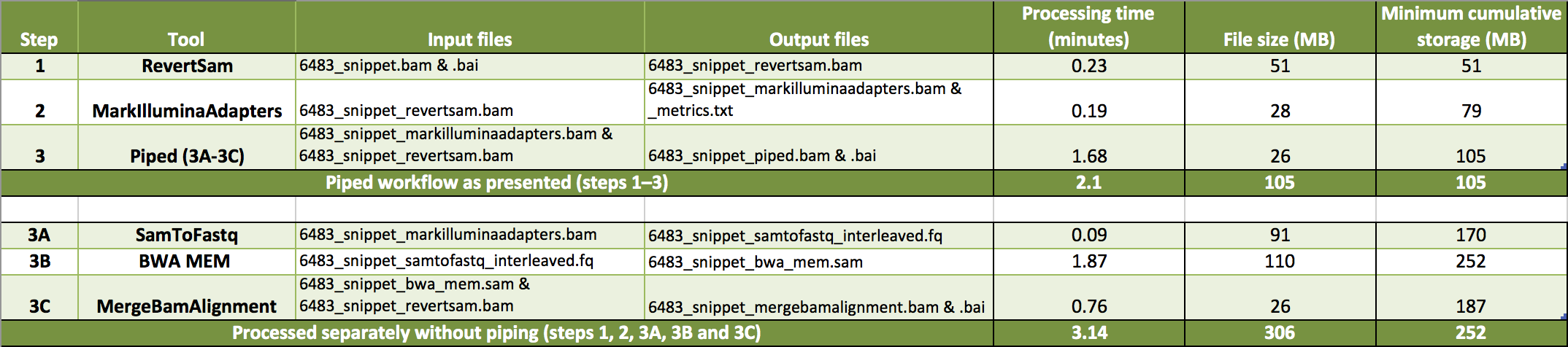
samtools index with cram: EOF marker is absent. The input is probably truncated. · Issue #146 · samtools/samtools · GitHub
![Error in alignment/'[W::bam_hdr_read] EOF marker is absent. The input is probably truncated' · Issue #439 · FelixKrueger/Bismark · GitHub Error in alignment/'[W::bam_hdr_read] EOF marker is absent. The input is probably truncated' · Issue #439 · FelixKrueger/Bismark · GitHub](https://user-images.githubusercontent.com/64626735/120408365-e2698980-c31c-11eb-834b-66cde1d95b69.png)
Error in alignment/'[W::bam_hdr_read] EOF marker is absent. The input is probably truncated' · Issue #439 · FelixKrueger/Bismark · GitHub
split by reference chr = "EOF marker is absent. The input is probably truncated." · Issue #137 · pezmaster31/bamtools · GitHub

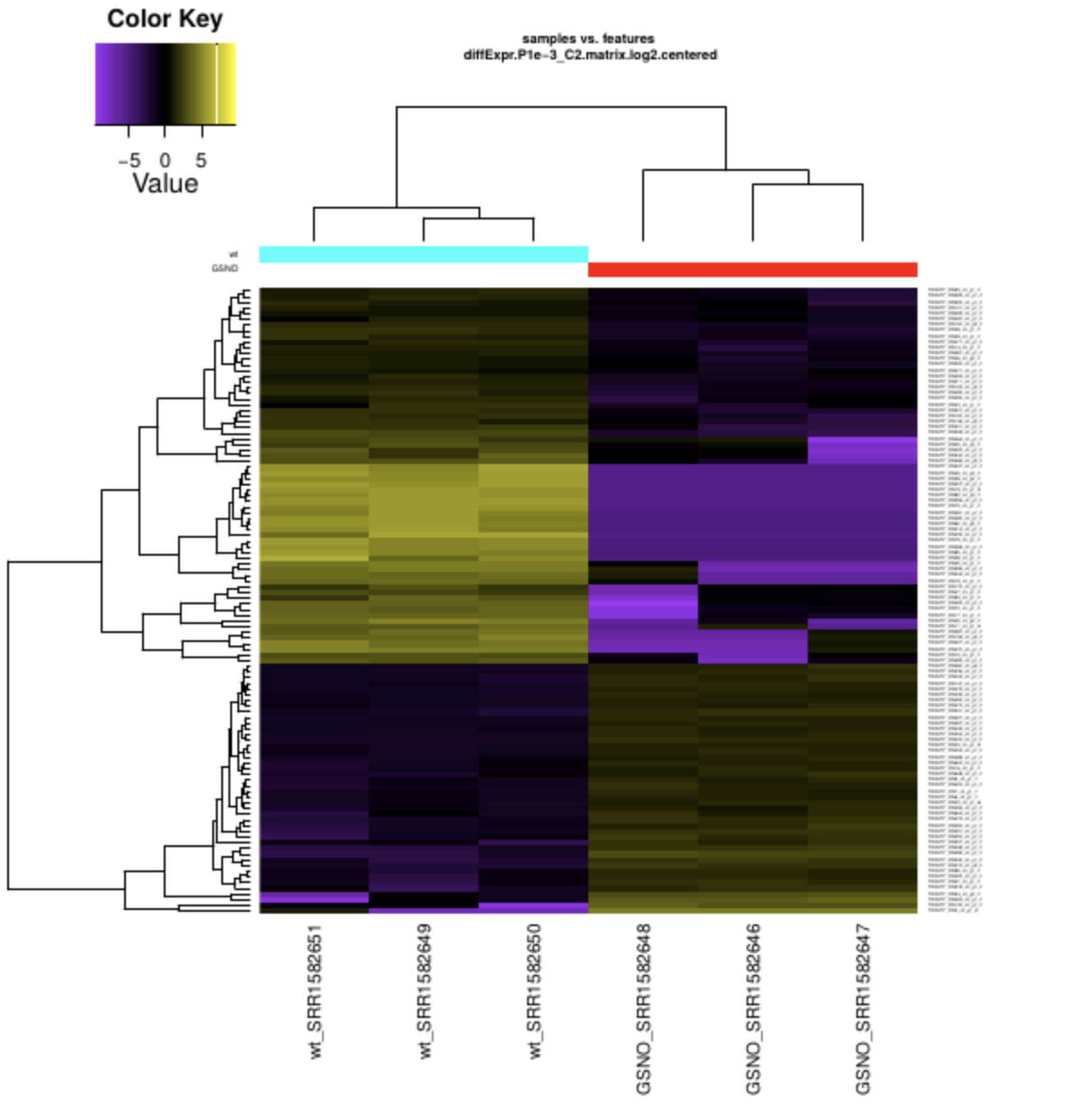
RiboProfiling: a Bioconductor package for standard Ribo-seq pipeline processing. - Abstract - Europe PMC
W::bam_hdr_read] EOF marker is absent. The input is probably truncated. · Issue #382 · freebayes/freebayes · GitHub
W::bam_hdr_read] EOF marker is absent. The input is probably truncated. · Issue #382 · freebayes/freebayes · GitHub

RiboProfiling: a Bioconductor package for standard Ribo-seq pipeline processing. - Abstract - Europe PMC

RiboProfiling: a Bioconductor package for standard Ribo-seq pipeline processing. - Abstract - Europe PMC
![Error in alignment/'[W::bam_hdr_read] EOF marker is absent. The input is probably truncated' · Issue #439 · FelixKrueger/Bismark · GitHub Error in alignment/'[W::bam_hdr_read] EOF marker is absent. The input is probably truncated' · Issue #439 · FelixKrueger/Bismark · GitHub](https://user-images.githubusercontent.com/64626735/122110946-26649000-cded-11eb-884b-4281fa2b161f.png)